COVID-19 SIRf Modelling
Implementation of Oxford’s Susceptible-Infectious-Recovered framework (SIRf) Coronavirus (CV19) tracking model. Uses SciPy’s ODE integrator to integrate a set of coupled linear first order ODEs with adjustable population parameters. Includes option to modify the effective reproductive number (‘R’) of the virus to see the effect of lock-down measures.
Sample Output
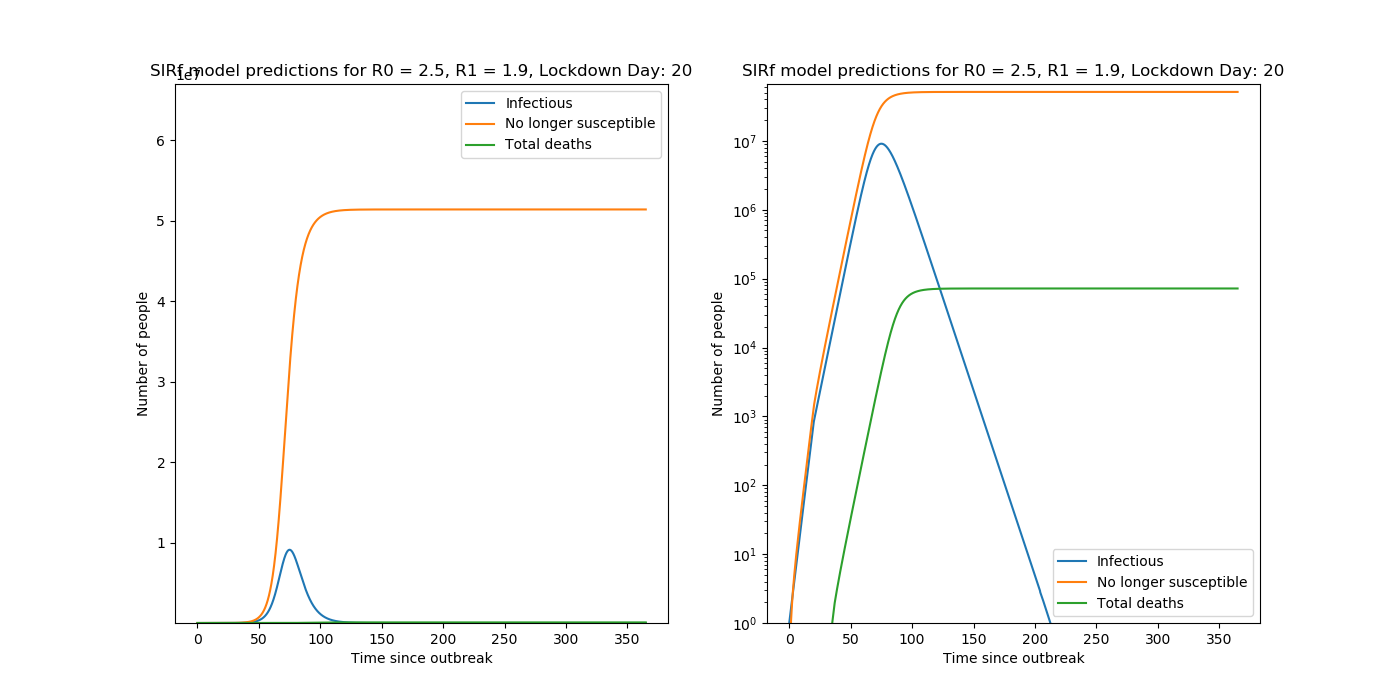
Predictions are made with the following set of coupled ODEs,
\[\frac{dy}{dz} = by(1-z) - \frac{y}{T}\] \[\frac{dz}{dt} = by(1-z)\] \[D = NPaz(t-1)\]where \(b = \frac{R}{T}\) .
These are integrated with the SciPi ODE Integrator:
# Integrate the model with SciPi ODE Integrator
xt = odeint(SIRf, x0, t, args=(b, T))
SIR Model
Parameters:
- y(t) Proportion of population who are infectious
- z(t) Proportion of initial population no longer susceptible to infection (dead, infected or recovered)
- D(t) Total deaths
- t Time since outbreak began
- R Basic reproduction number
- T Average infectious period
- b Average number of people infected by an infectious individual per day (R/T)
- l Average time between infection and death
- a Probability of dying with severe disease
- p Proportion of population at risk of severe disease
- N Size of population
See source code on my GitHub for parameter values used.

